4 Dataset provided for download
4.1 The details of four Dataset
We provide downloads of variant datasets, including single nucleotide polymorphism (SNP), insert and deletion (InDel, with size ≤50bps), and large structural (SV, with size ≥51bps) variant datasets (Figure4.1). The SNPs and small InDels are identified among all 2,839 rice hybrids and 486 parental lines of hybrids. Variants are called using “HaplotypeCaller”, “GenomicsDBImport” and “GenotypeGVCFs” functions in GATK (the genome analysis toolkit v4.1.4.1) with default parameters (McKenna et al., 2010). Variant filtration is conducted using “VariantFiltration” function in GATK with parameters of “–cluster-size 3 –cluster-window-size 10 QD<10.00 FS>15.000 AC<3 DP>200||DP<5” for SNPs and “QD<10.00 FS>30.000 DP>200||DP<5” for InDels. And the SVs are identified among 964 rice hybrids. SVs are identified using an graph-based genome (Qin et al., 2021) and an SV genotyping pipeline integrated in Variation graph toolkit (Garrison et al., 2018).
Figure 4.1: SNP, InDel and SV variant datasets for download
In addition, a differentiated indica-japonica variant dataset is also available (Figure4.2). This dataset comprises 830,245 SNPs, which are identified according to the following criteria:
at an indica-japonica differentiated SNP site,
≥17 indica varieties are the same genotype;
≥21 japonica varieties held the same genotype;
Indica and japonica rice accessions possess different genotypes.
Figure 4.2: Differentiated indica-japonica variant dataset for download
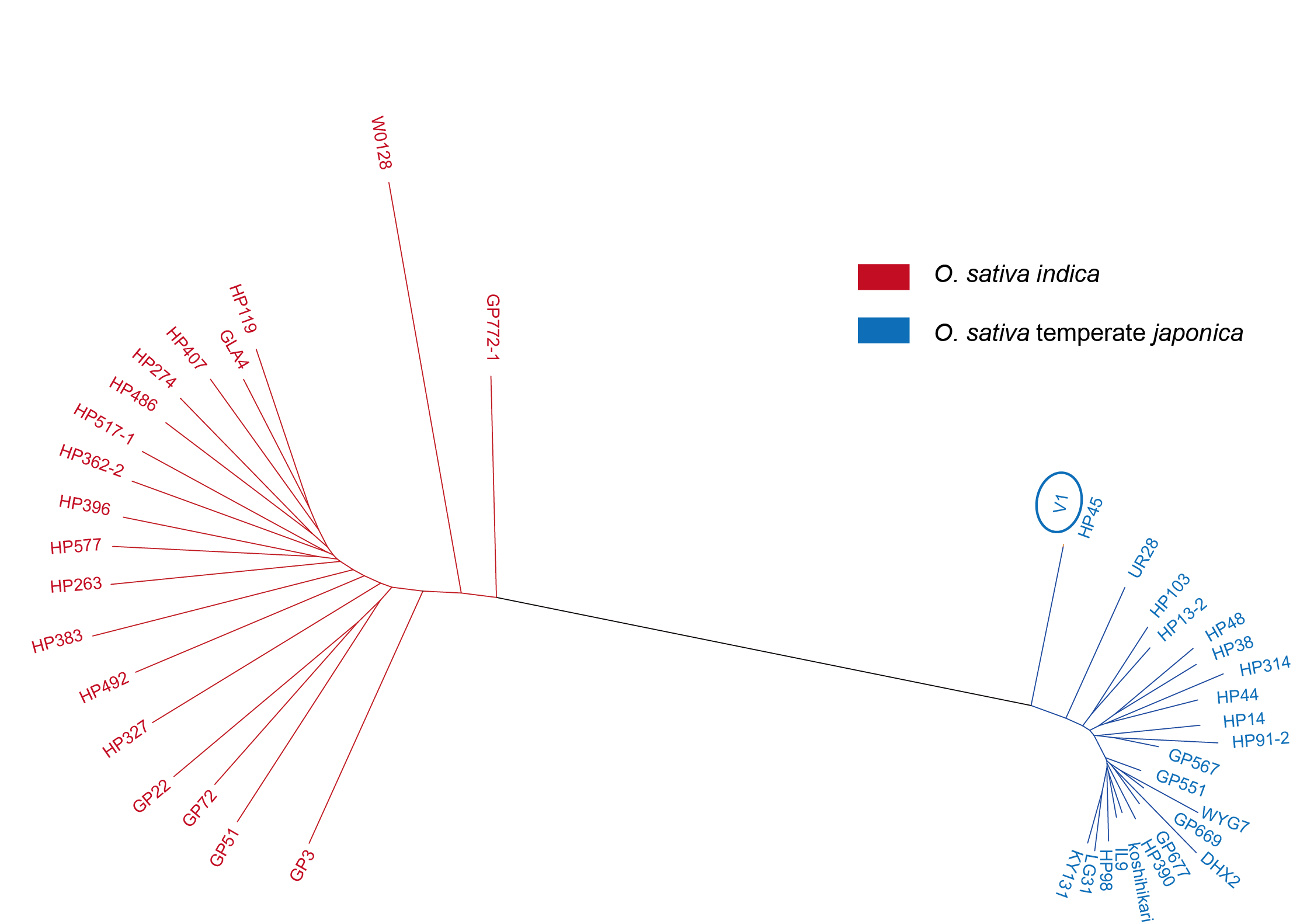
Figure 4.3: Neighbor-joining tree of 42 rice accessions used for differentiated SNPs identification
4.2 Reference
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297-1303 (2010).
Qin, P. et al. Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell 184, 3542-3558 (2021).
Garrison, E. et al. Variation graph toolkit improves read mapping by representing genetic variation in the reference. Nat. Biotechnol 36, 875-879 (2018).
Zhao, Q. et al. Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat Genet 50, 278-284 (2018).