1 Home search genes by IDs, functions, and pathways
On the index page, a Home search function is provided for quickly accessing one or a series of genes. Normally, we may not remember the IDs of genes. But, we could search genes by function descriptions or keywords. This would facilitate the usage of VPI-MD.
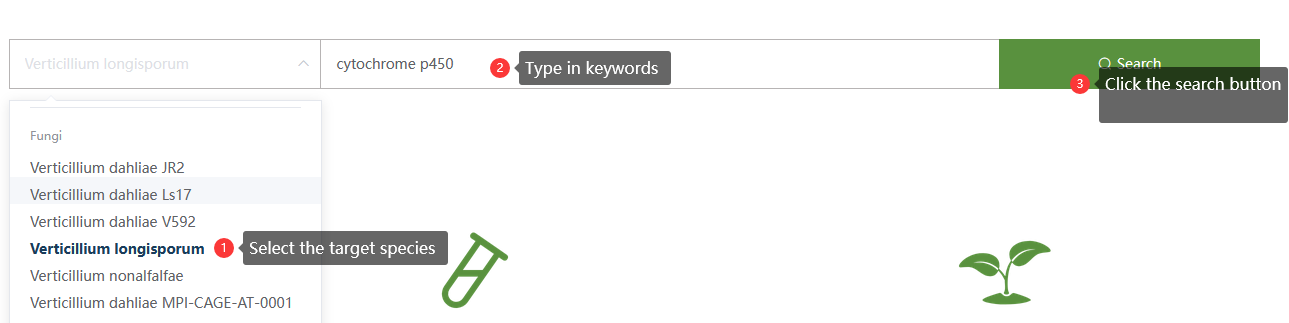
For example, we want to search for cytochrome p450 genes in Verticillium longisporum. We could operate in the following 3 steps:
Totally, 114 related genes are returned. From the descriptions, we could see some genes are Cytochrome P450 (these are what we searched).
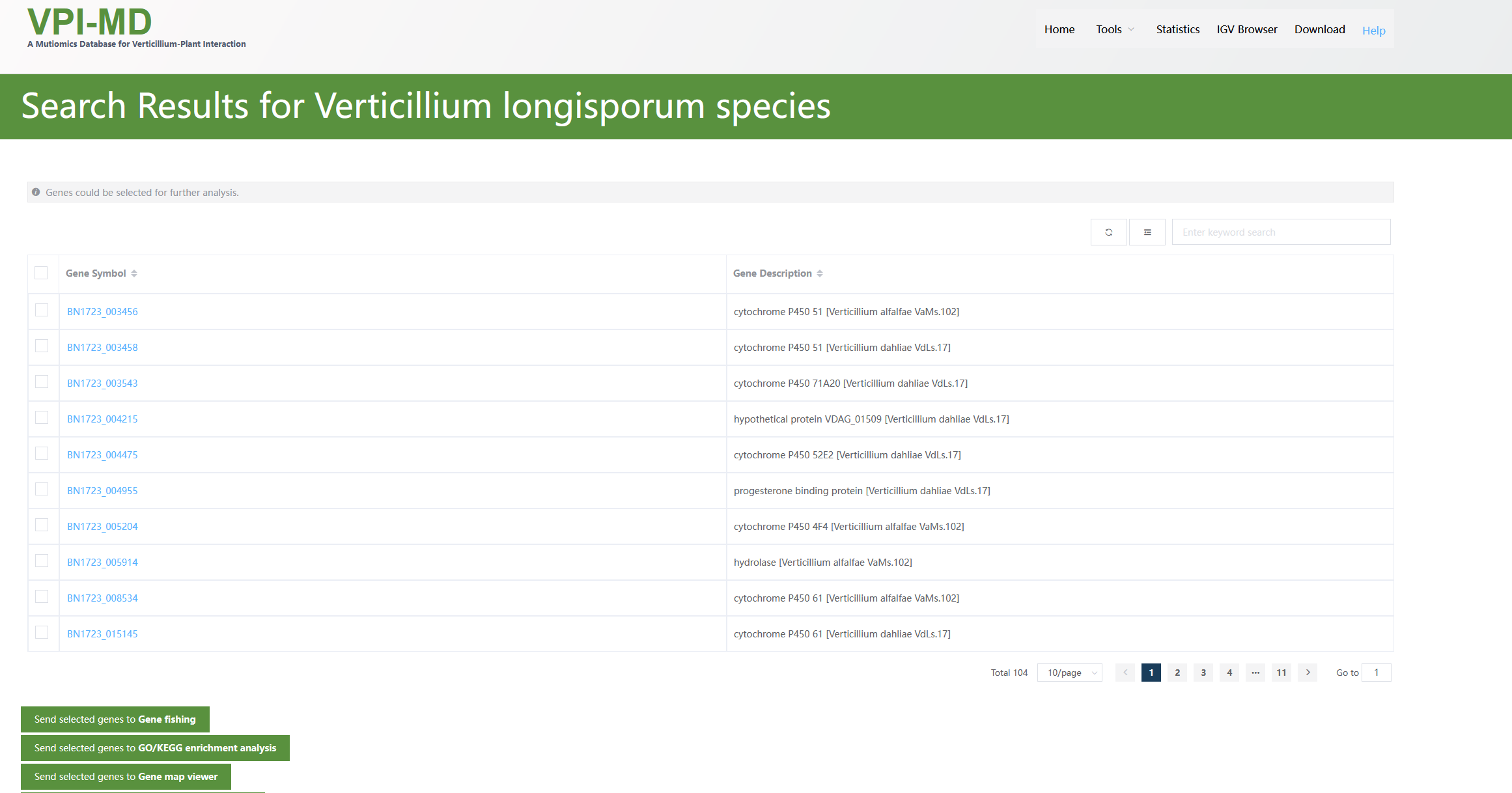
The annotations of some genes may not correspond to Cytochrome P450. This is not wrong since the keyword Cytochrome P450 must be contained in one of their functional annotations. We could check the detailed information of these genes by clicking the gene names.
1.1 Gene detail page
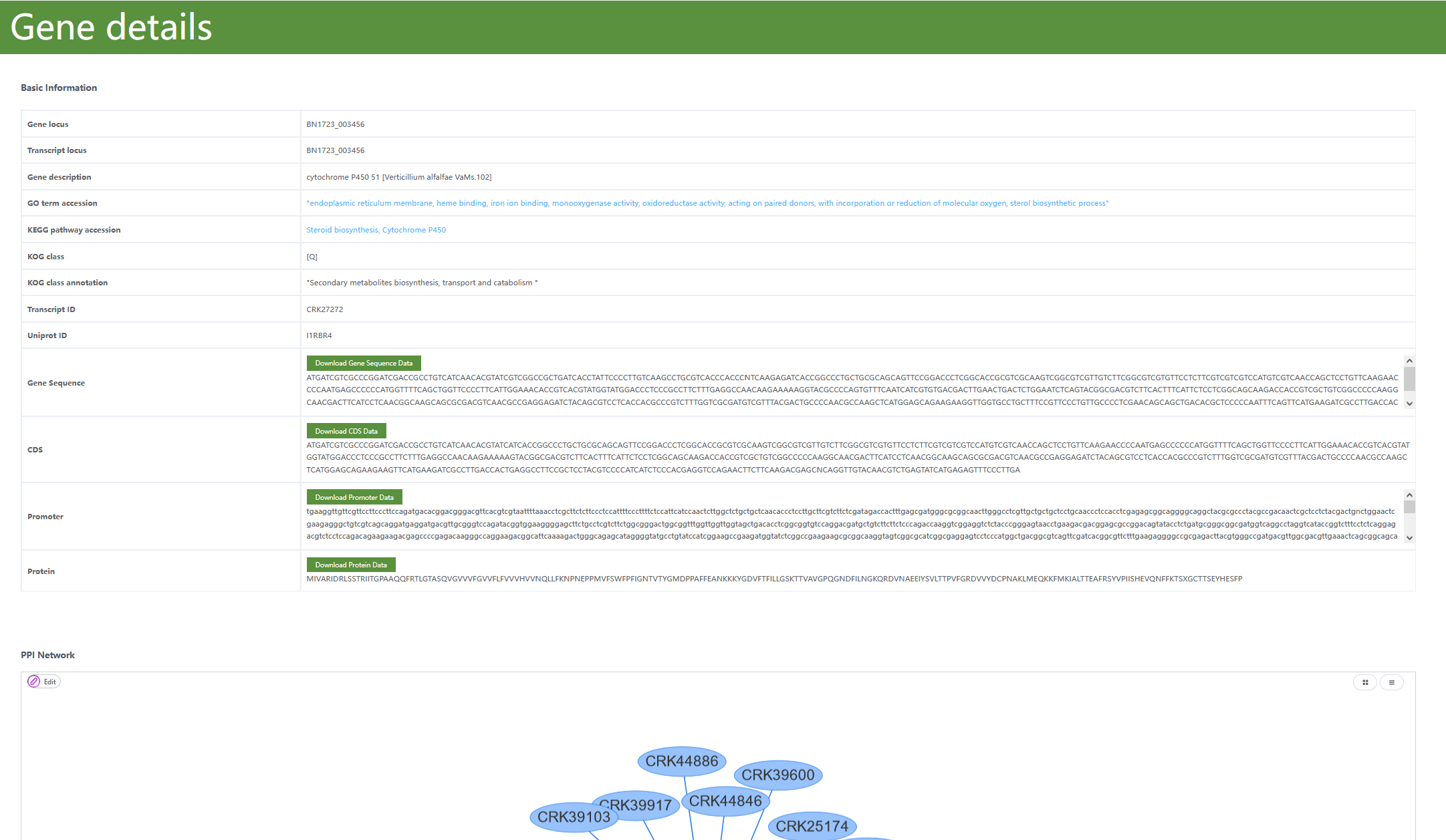
As for transcript BN1723_003456, this is a cytochrome P450 51 gene. The below screenshot could be classified into 3 parts.
The basic annotation information like
Gene locus,Transcript locus,Gene description,GO term name,KEGG Pathway accession,KOG class,KOG class annotation,Transcript ID,Uniprot ID. One could click the blue text to tour Gene ontology, and the KEGG database for more information. Normally, these page transitions are not required.The sequence part contains
Gene,CDS,Promoter(upstream 2 kilobases of the transcription start site as defined as promoters here), andProteinsequences of the gene. These sequences could be directly copied or downloaded for other usages.PPI Network: one can identify potential interacting genes that may engage in biological interplay with the focal gene. This information is derived from the STRING database, a well-established repository that integrates empirical data and computational predictions to delineate the interactome landscape of genes within the organism under study.