6 New DE Gene analysis
This portion of the system operates similarly to the ‘DE Gene analysis’ module, primarily designed to process custom data uploaded by users, which may not be present in the existing database.
Using limma-voom to screen DE genes based on Read-count expression data and performing GO/KEGG enrichment analysis for differentially Expressed (DE) genes.
6.1 First step: Upload data and parameter selection
Just select each parameter sequentially as labeled in the figure.
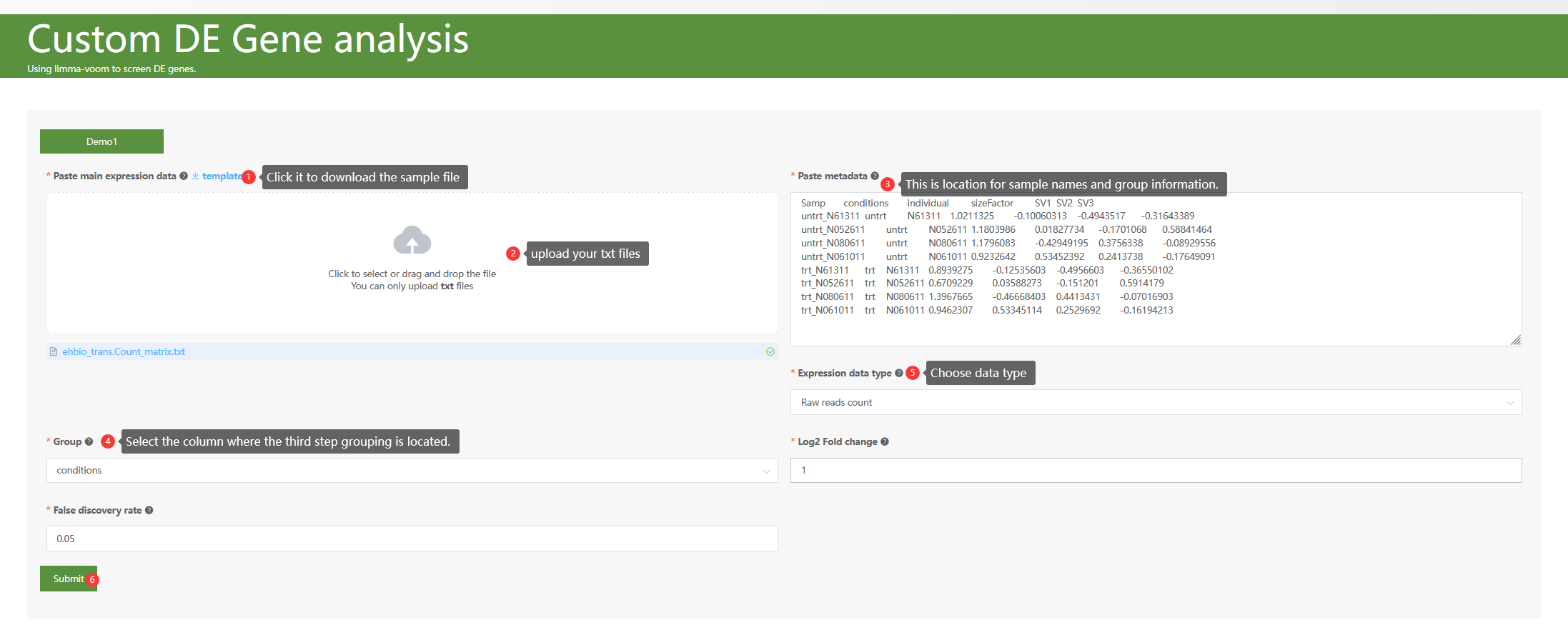
Figure 6.1: A screenshot of the first step of DE gene analysis.